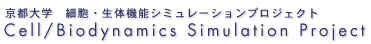Run
Start simBio_JavaSpaces
The start order for the system is shown below.
- A protocol XML file for sensitivity analysis is processed on a client alone to confirm the efficacy. Duration and repeat times of calculation can be decreased at this stage.
- A JavaSpaces server is started.
- Workers are started.
- An http class server is started.
- The protocol XML is executed with an option to use distributed computing.
- Graphs will be presented right after computation.
Client, and each server of Javaspaces, simBio, and http can be run on separate computers or on the same PC. The client and the http server are typically executed on the same PC.
Start RemoteClient
If the RemoteClient will be used start it by following the instruction, Start RemoteClient.
Start simBio_JavaSpaces servers
To start the simBio_JavaSpaces servers run the DOS batch file startJavaSpaces.bat or the UNIX/LINUX Shell Script startJavaSpaces.sh depending on which operating systems the server is running on.
After a short time a window with similar look as the figure shown below will appear. A cursor will flash on the next line.
Start simBio_JavaSpaces workers
simBio_JavaSpaces workers can be started in two different way. One which require that the user visit all computers acting as workers and another that makes it possible to start all workers in one single command.
Start simBio_JavaSpaces workers Local
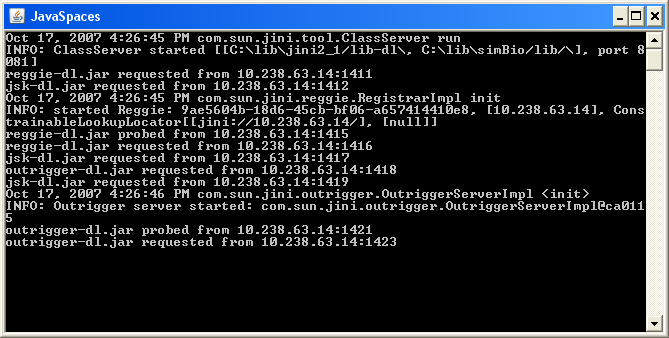
To start the simBio_JavaSpaces workers local, run the DOS batch file startWorker.bat or the UNIX/LINUX Shell Script startWorker.sh depending on which operating systems the workers are running on.
After a short time blue windows with similar look as the figure shown below will appear. A cursor will flash on the next line.
Start simBio_JavaSpaces workers Remote
To start the simBio_JavaSpaces workers remote use the RemoteClient's start command.
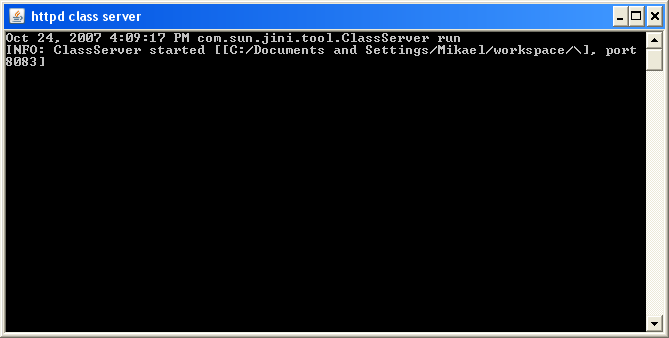
Start http server
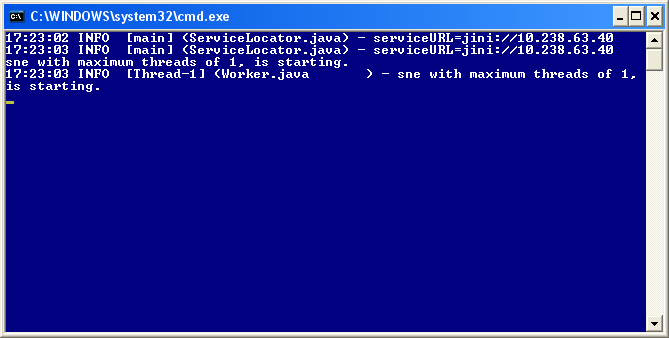
To start the http server, run the DOS batch file httpd.bat or the UNIX/LINUX Shell Script httpd.sh depending on which operating systems the http server is running on.
After a short time a window with similar look as the figure shown below will appear. A cursor will flash on the next line.
Run model in simBio_JavaSpaces
Now when everything are up and running it is time to run a model in simBio_JavaSpaces. For this example, we will use the model NCXvsCaL found in the simBio project at src/xml/sarai_et_al_2006/figure/3A_NCXvsCaL.xml.
First verify that the protocol XML-file has been adjusted for the system. Change the ServiceURL and Codebase so it fits your system.
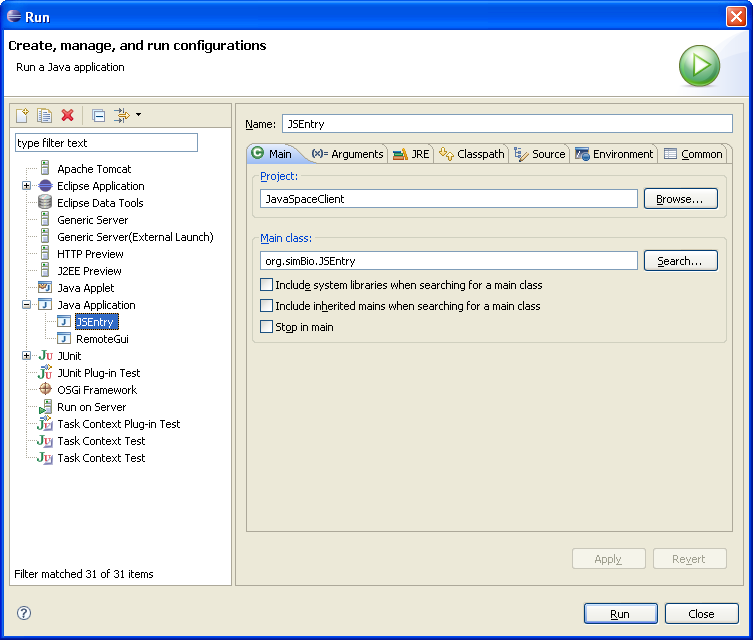
To run the model, choose the menu [Run] -> [Open Run Dialog...] in Eclipse. In the run dialog choose to create a [New launch configuration] of the type [Java Application] and give the new configuration a name.
In the Main tab set Project to JavaSpacesClient and the Main class to org.simBio.JSEntry as the figure below shows. The settings may be different depending on Eclipse version. This is how it is done in Eclipse Europa (3.3).
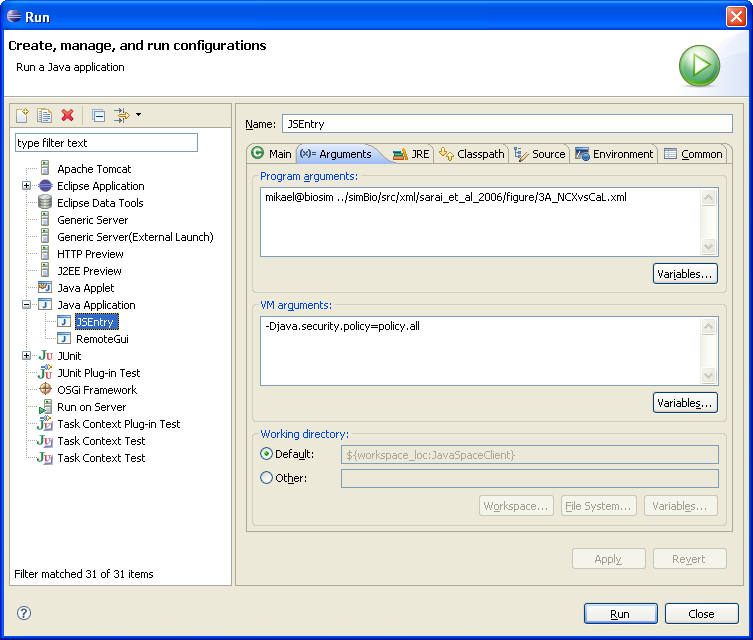
In the Arguments Tab set Program arguments to your e-mail followed by model path and VM arguments to -Djava.security.policy=policy.all as the figure below show. The settings may be different depending on Eclipse version. This is how it is done in Eclipse Europa (3.3).
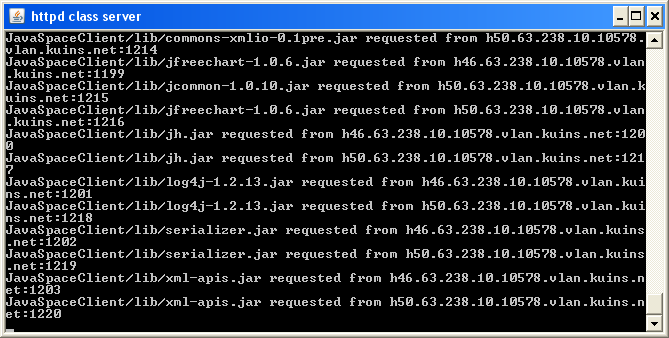
The simBio_JavaSpaces workers will now use the Codebase to search for the code to execute. The http server will log this in a similar way as the figure shown below.
During the calculation simBio_JavaSpacesClient will collect calculated XML-files from the workers in the folder baseDir\xmlDir. When the calculation finish a CSV-file and a Toc-file will be generated according to the tags baseDir\csvFile and baseDir\xmlFile. Finally a plot will appear which use the settings that are read from the settings attribute for the evaluated variables. See generate and evaluate for more information.
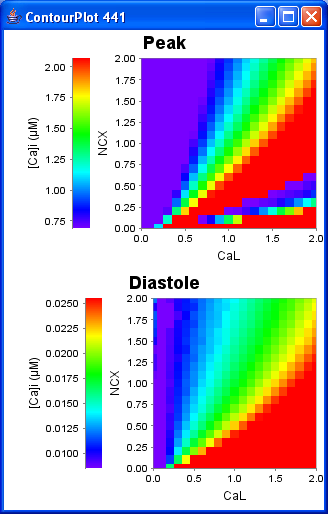
The figure below show the expected contour plot for Sarai et al 2006 Fig 3A (NCXvsCaL).
Run model in simBio_JavaSpaces from the simBio binary release package
If the binary release package of simBio is used then must Project in figure 5 (Run - Main Tab) be set to simBio. The box Include system libraries when searching for a main class must also be checked to find the Main class org.simBio.JSEntry.
For the example above, the path for the model NCXvsCaL is found in the simBio project at xml/sarai_et_al_2006/figure/3A_NCXvsCaL.xml. Therefore in figure 6 (Run - Arguments Tab), Program arguments for the model path should be set to xml/sarai_et_al_2006/figure/3A_NCXvsCaL.xml.
Run model in simBio_JavaSpaces with the simBio Eclipse plug-in
If the binary release package of simBio is used or if a private simBio project is used then it is possible to start executing a model in simBio_JavaSpaces by first selecting a XML-file and then open the pop-up menu [simBio]->[Run model on JavaSpaces].
The default e-mail address that will be used is email@address.com. This can however be configured by creating a file with the name simBio-plugin.properties in the rootpath of the running project. The file should contain the key EMAIL, as shown in the example below.
EMAIL=user@sim-bio.org
Shutdown simBio_JavaSpaces
The system has no shutdown order.
Shutdown simBio_JavaSpaces workers
simBio_JavaSpaces workers can be shutdown in two different way. One which require the user to visit all computers acting as workers and another that makes it possible to shutdown all workers in one single command.
Shutdown simBio_JavaSpaces workers Local
To shutdown the simBio_JavaSpaces workers locally run the DOS batch file stopWorker.bat or the UNIX/LINUX Shell Script stopWorker.sh depending on which operating systems the workers are running on.
A green window with similar look as the figure below will appear and after a short time both the green and blue windows will disappear.
Shutdown simBio_JavaSpaces workers Remote
To shutdown the simBio_JavaSpaces workers remote use the RemoteClient's shutdown command.
Shutdown simBio_JavaSpaces servers
To shutdown the simBio_JavaSpaces servers, press Ctrl+c when the window is active.
Shutdown http server
To shutdown the http server press Ctrl+c when the window is active.
Shutdown RemoteClient
If the RemoteClient was used, shut it down by following the instruction, RemoteClient's exit command.
Start using simBio_JavaSpaces
Now it is time to create a private project and start using simBio_JavaSpaces, follow the instruction Create a private project to do this, enjoy.