II-2 Changing Parameters from XML
In simBio, calculation formulas of functional elements are described in Java format, and the initial value of parameters and the model structure are described in XML format.
About XML
XML is one way of expressing data in a text format and is widely used as a technique of exchanging data between programs, for example used for the description of web pages.
The XML document of matsuoka_et_al_2003/model.xml is structured as followings.
First of all, in the first line, it is specified that this text file is in the XML version 1.0 format, and is encoded in UTF-8.
In the second line there is a comment written between <!-- and --> .
This XML document describes data like the following.
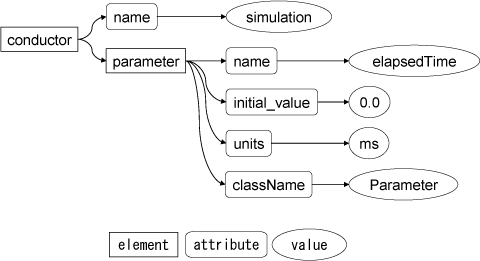
The basic information unit of an XML document is called an "element", in lines 3-5 there is a conductor element which has child elements, and in line 4 there is a parameter element which do not have child elements.
In XML, data is described in nested structure. The starting tags are strings of the element name which are enclosed between < and >, e.g conductor. The ending tags are strings which are enclosed between </ and > and between those tags the child elements are described. Therefore, the parameter element between the beginning and end tags on lines 3 and 5, is the child element of conductor.
"name", which follows the beginning tag, is called an "attribute", which provides more detailed information, and simulation is a "attribute value". In the program, simulation is set as the attribute value of the name attribute of the conductor element. Between the attribute and the attribute value are connected with =, and the attribute value is enclosed with ".
An empty element which does not have a child element is enclosed by < and />. In line 4, the parameter tag does not have child elements. The className attribute is set to "org.simBio.core.Parameter", the name attribute to "elapsedTime", the initial_value attribute to "0.0", and the units attribute to "ms".
A data structure which connects multiple child elements to an element in this way is called a tree structure, and an element of that tree structure without a parent is called the root. An XML document describes tree-structured data with text, and programs interpret the text as tree-structured data.
Interpreting of XML by simBio
As was mentioned earlier, the calculation formulas of the mathematical models in simBio are described using Java, and the initial values and ways of combining them are described in XML.
The XML document in Figure 2-2-1 is transformed into the instance shown in Figure 2-2-3.
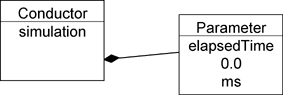
simBio recognizes the name, initial_value and units properties as written above, as well as className. In simBio, 1 Java class is described for 1 functional element, and an XML tag is connected to a class, by writing a fully qualified class name in the className attribute value.
Therefore simBio creates 1 instance of a related class for one element of an XML document, and creates a tree-structured instance in memory with a 1:1 correspondence to the XML document. In simBio, a class that displays functional elements is called Reactor, and its parameter is called Node, so when a model XML is read in, a Reactor and Node tree is completed. A Reactor can have Nodes so corresponds to a folder in a file system, and a Node to a file in a folder.
simBio distinguishes the elements of the model by name, so the name attribute must be set. Also, the className attribute must be set for the tags when they first appear in the document, but after that it is possible to be omitted. The initial value and units are set when required.
Changing the model XML
The model parameters which are displayed in the simBio GUI correspond 1:1 to the model XML document, so you can change parameter values by directly editing the XML file. Moreover, by editing the XML document you can add and delete features of specific ion channels and transporters. Here we shall explain about the editing of XML with Eclipse, using a pacemaker cell model as an example.
To freely edit an XML file, first create your own Java project, then copy and edit an XML file from simBio project.
Creating your own project
- Click on [File]->[New]->[Project] in the Eclipse menu likewise Section I-2 Settings for using source packages
- In the New Project dialog, select [Java Project], and click on [Next].
- Enter a suitable name in the project name box (Here enter tutorial), and when you click on [Finish], the tutorial project will be created.
- To utilize the functions of simBio, right click on tutorial and click on [Properties].
- Select [Java Build Path], open the [Projects] tab and click on [Add].
- In the Required Project Selection dialog put a check in simBio, and when you click on [OK], the simBio project will
be added to the build path.
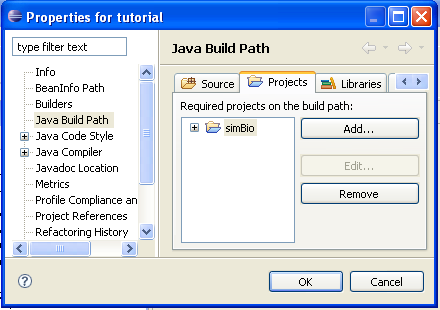
- Click on [OK], and include the simBio project in the build path.
XML Duplication
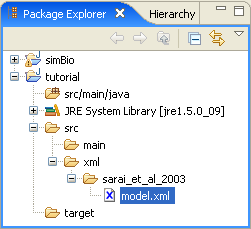
- Right click on the src folder of the tutorial project and click on [New]->[Folder], enter xml in [Folder Name] and create the xml folder.
- Right click on the simBio project folder src/xml/sarai_et_al_2003 and copy.
- When you right click on the tutorial/src/xml folder which you created and select Paste, sarai_et_al_2003 will be
duplicated.
 Keyboard shortcutYou can edit more quickly by using keyboard shortcuts than by using the mouse. In the case of Windows, you can copy a folder by selecting it and then pressing the [Ctrl] and the [C] key at the same time. To paste, press the [Ctrl] and the [V] key at the same time.
Keyboard shortcutYou can edit more quickly by using keyboard shortcuts than by using the mouse. In the case of Windows, you can copy a folder by selecting it and then pressing the [Ctrl] and the [C] key at the same time. To paste, press the [Ctrl] and the [V] key at the same time.
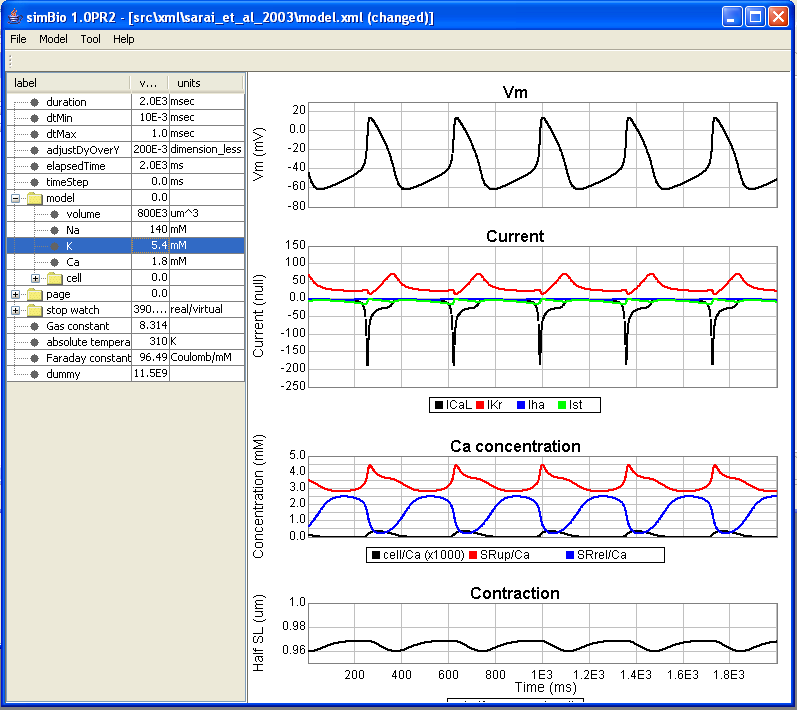
Changing the K Concentration
Double click on tutorial/src/xml/sarai_et_al_2003/model.xml and open it. The extracellular K+ concentration is set in simulation/model/K, so change it from 5.4 mM to 15.0 mM.
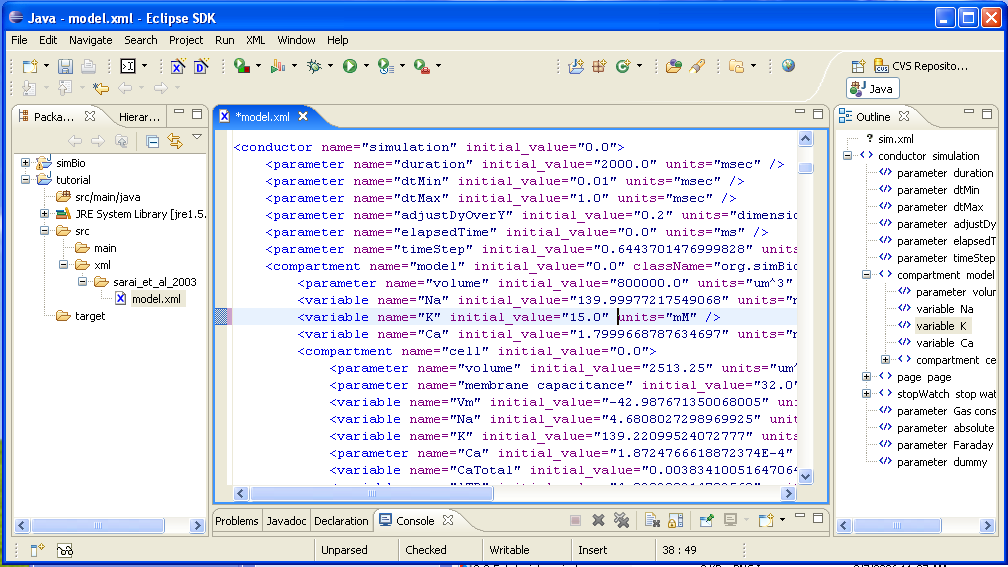
Click on [File]->[Save] in the menu, and save your changes.
Right click on model.xml, and click on [simBio]->[Run on GUI] to open the GUI.
When you open the [model] parameter, you can see that the extracellular K+ concentration has been changed from 5.4 mM to 15.0 mM. When you run it in this state, you can likewise run the simulation with the changed parameters.
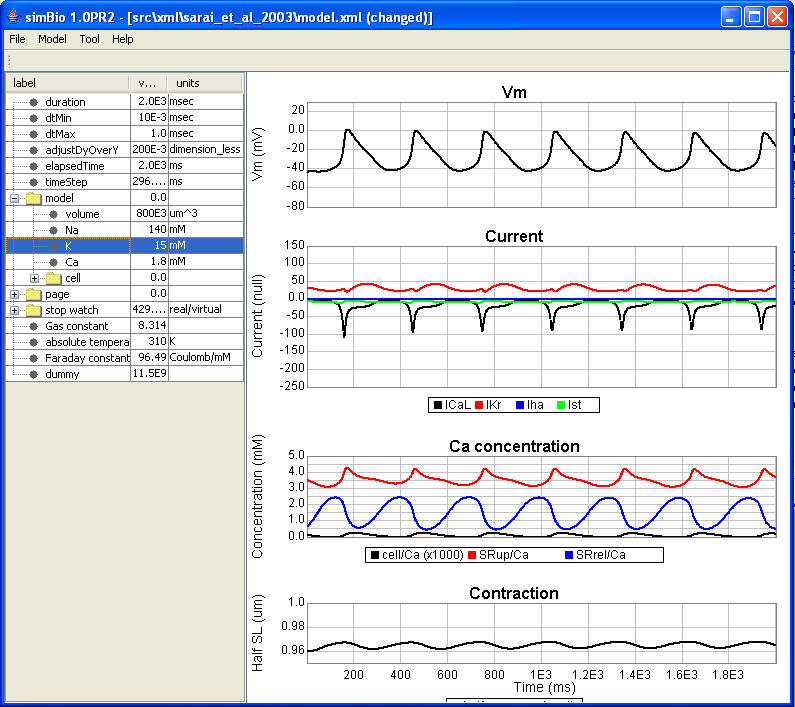
The result is that, when the extracellular K + concentration of 5.4 mM is compared with the 15.0 mM case, it can be seen that the resting membrane potential becomes shallow, and the degree of depolarization also becomes smaller.